Optical Nanoscopies and Biophotonics
New superresolution techniques to study the fast spatiotemporal orchestration of proteins in cell adhesion and in neurons.
By delivering optical images at high rates and with spatial resolutions below the diffraction limit, super-resolution fluorescence microscopy has opened new opportunities to study biological structures with finer details such as the fast spatiotemporal orchestration of proteins in cell adhesion and in neurons.
Achievements and Projects
Massive parallelization of STED nanoscopy
One of the great challenges for Super-resolution microscopies is to achieve fast wide-field imaging while maintaining the nanoscale super-resolution. STimulated Emission Depletion (STED) nanoscopy stands out by instantly providing signals from predetermined nanosized regions in the sample and can in principle provide fast imaging speeds. However, STED nanoscopy has long remained a point-scanning technique and needs parallelization.
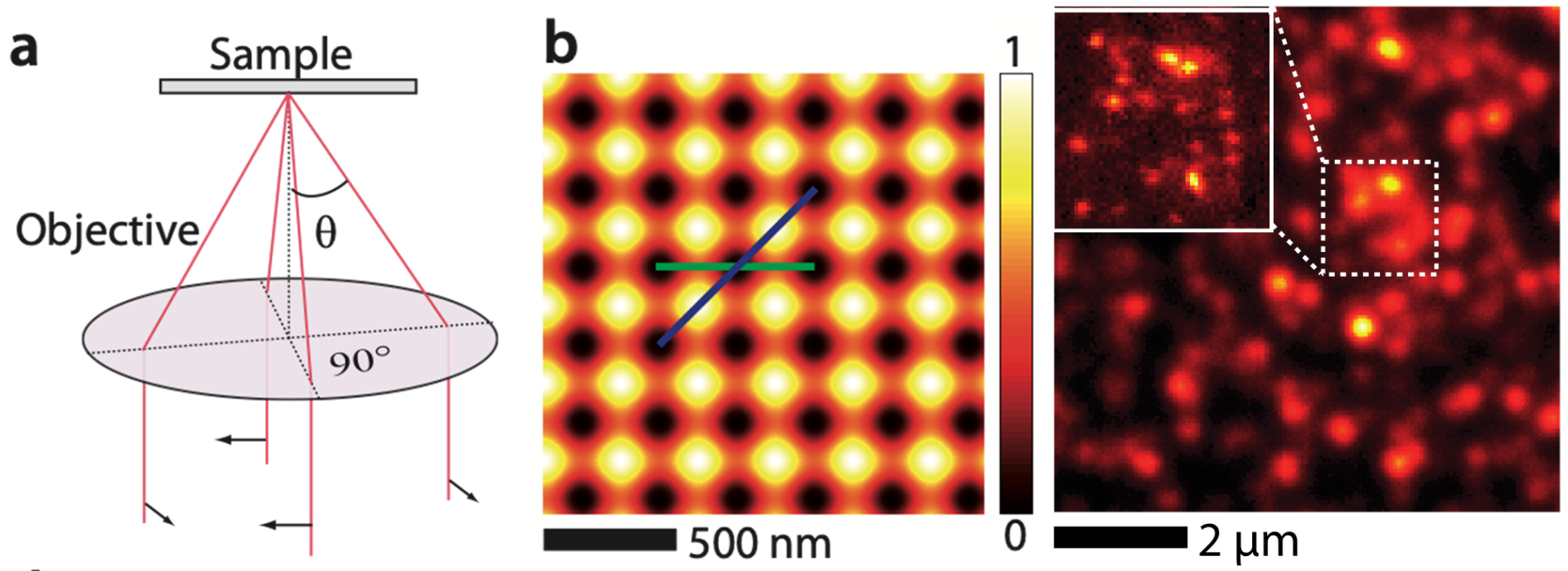
We have demonstrated massive parallelization of STED nanoscopy using wide-field excitation together with well-designed optical lattices for depletion and a fast camera for detection. Acquisition of super-resolved images requires scanning over a single unit cell of the optical lattice, which can be as small as 300nm x 300 nm for a total scanning time of ~ 80 ms per image. Optical lattices achieve efficient fluorescence depletion with moderate laser powers (Optics Express, 22, 5, 5581-5589 (2014)) over fields of view as large as ~8 µm x 8 µm. This approach opens many perspectives in fast wide-field super-resolution imaging, such as the study of endogenous proteins in living cells.
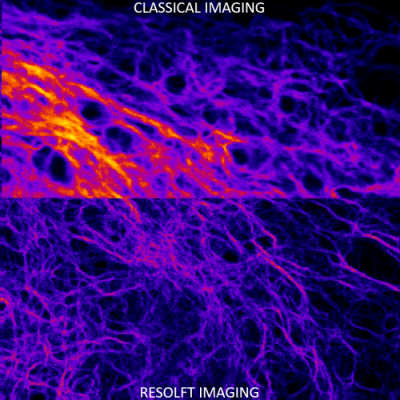
Revealing the nanoscale dynamics of cell focal adhesions using RESOLFT nanoscopy
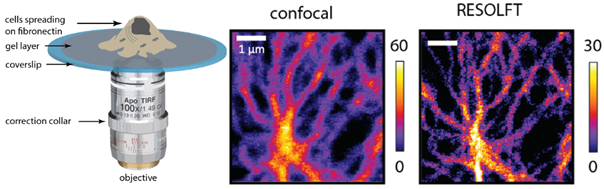
Cells can adjust their adhesive and cytoskeletal organizations, depending on the changes in the biochemical and physical nature of their surroundings. Nanoscale protein complexes called focal adhesions are the integrating points of these regulating signals. RESOLFT (REversible Saturable OpticaL Fluorescent Transitions) is a nanoscopic imaging technique that overcomes the diffraction barrier by suppressing the volume of emission through a reversible on-off process in reversibly switchable fluorescent proteins.
We use RESOLFT nanoscopy to study the protein dynamics in focal adhesions in vitro and on soft substrates with healthy migrating cells. To enable faster imaging, the RESOLFT setup was parallelized using optical lattices. A second lattice for fluorophore on switching has also been implemented to reduce the effects of photobleaching. The dual lattice setup enables long-term imaging (>30 frames) of nanostructures with resolutions close to 60 nm inside living cells. Such imaging can be performed at a frame rate of ~1Hz over a 15x15 μm2 field of view.
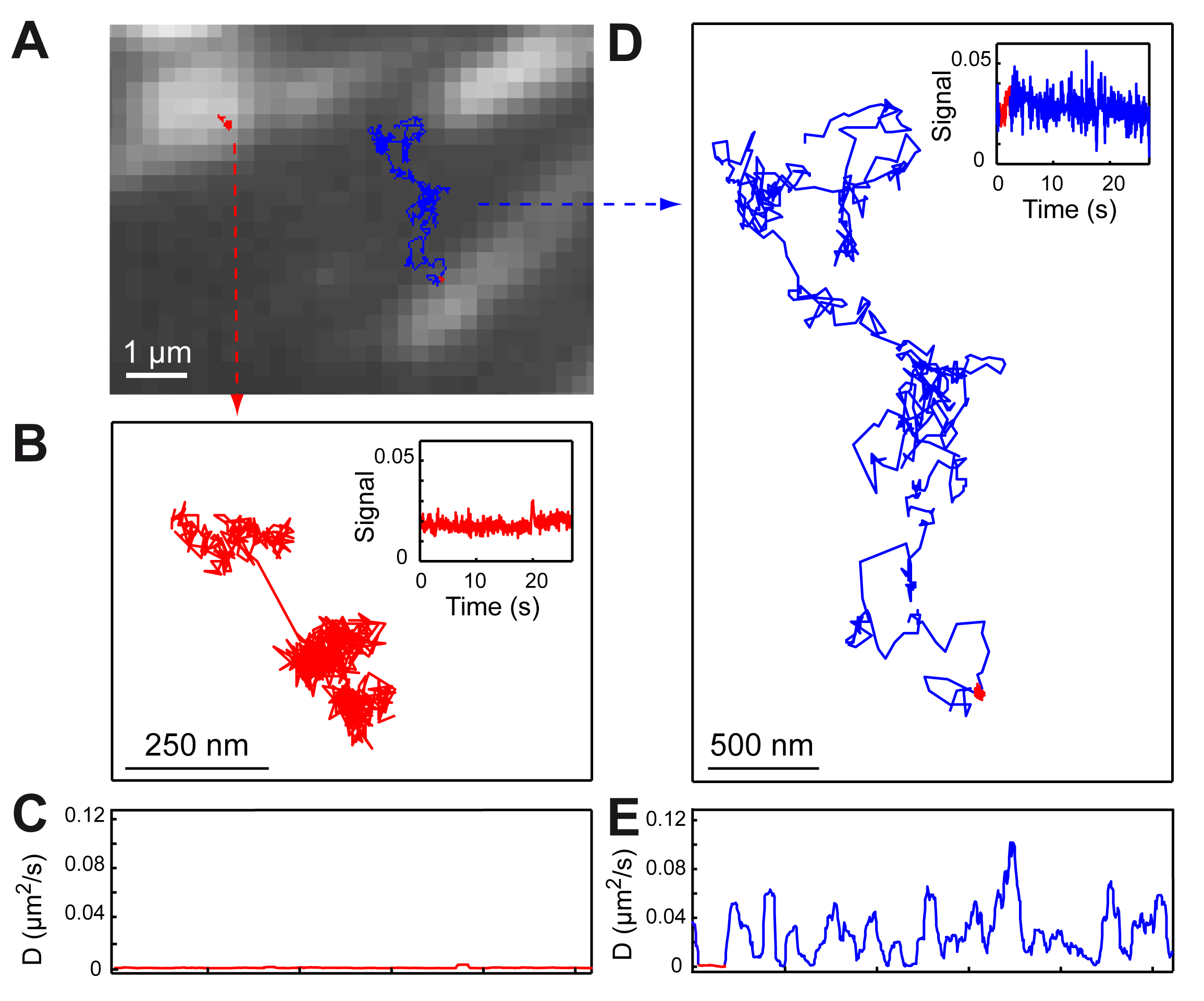
Photothermal microscopy
Single-molecule tracking in live cells is the ultimate tool to study subcellular protein dynamics, but it is often limited by the probe size and photostability. We have developed ultrasensitive optical techniques for the detection and study of non-fluorescing nano-objects (Science, 297, 5581, 1160-1163 (2002), Phys. Rev. Lett. 93, 257402 (2004)). These methods are based on the photothermal effect and have an unprecedented sensitivity allowing optical detection and absorption spectroscopy of tiny metal nanoparticles, non-luminescent quantum dots, and single-walled carbon nanotubes. We also have developed an innovative method for single nanoparticle tracking using triangulation (a “Nano-GPS”) that can record arbitrarily long particle trajectories. We have designed and characterized a full functional nano-sized probe for long-term single-molecule imaging: 5 nm gold nanoparticles functionalized with nanobodies, consisting in a small fragment of camelid antibody that recognizes widely used GFPs with a very high affinity (Nano Lett. 2013, 13, 4, 1489–1494 (2013)). These small and functional gold nanoparticles can be detected and tracked for unlimited periods of time in living cells using photothermal imaging. The versatility and targeting efficiency of our probes are demonstrated by labeling several types of GFP-tagged proteins, both in in vitro and cellular systems (Adv. Sci. 4, 1600280 (2017)). These probes can be used to study proteins not only in confined structures but also in intracellular compartments by tracking adhesive proteins and microtubule-associated proteins in living cells.